HOMCOS : Searching and Modeling of 3D Structures of Complexes
 Searching Contact Molecules for Query Protein
Searching Contact Molecules for Query Protein


 Searching Contact Molecules for Query Protein
Searching Contact Molecules for Query Protein
 Searching Contact Molecules for Query Protein
Searching Contact Molecules for Query Protein
This is the service for searching contact molecules with similar proteins to a given query sequence


| database | example |
| UniProt ID | CDK3_HUMAN [link] |
| UniProt AC | Q00526 [link] |
| INSDC protein_id | AAV40830.1 [link] |
| RefSeq protein_id | NP_001249.1 [link] |
Notes for input the query sequence
On the top of the windows, some common links and tables are shown among the three views.
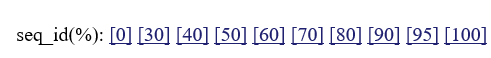 :if you click these numbers, you can change threshold sequence identity for choosing homologues.
:if you click these numbers, you can change threshold sequence identity for choosing homologues.
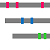
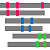 "Summary Bars" view
"Summary Bars" view
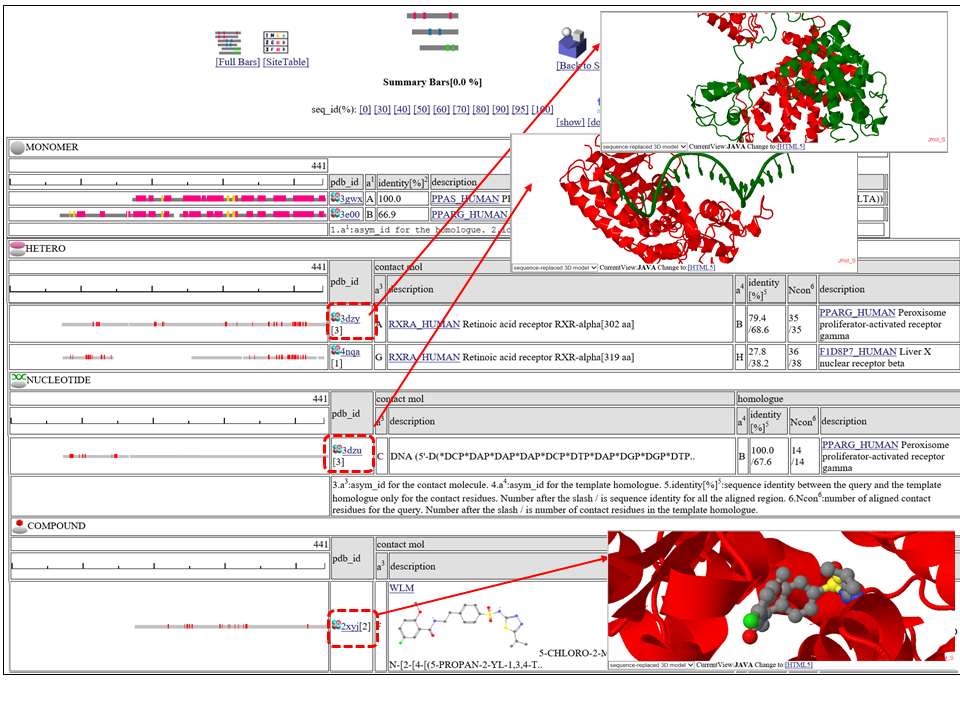

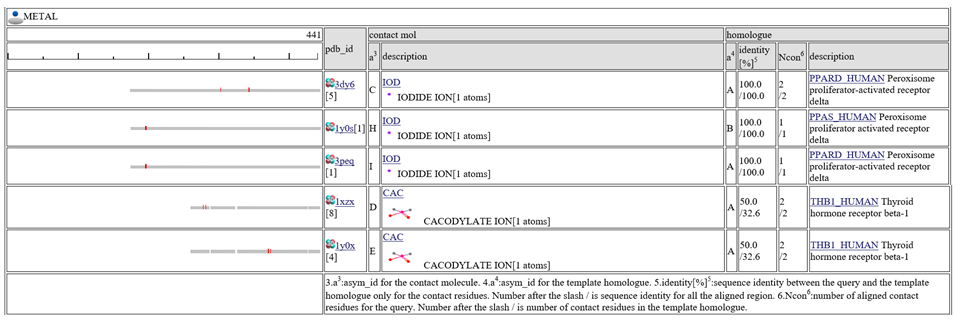
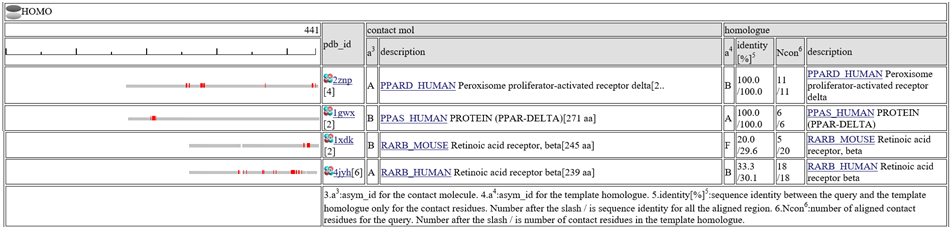
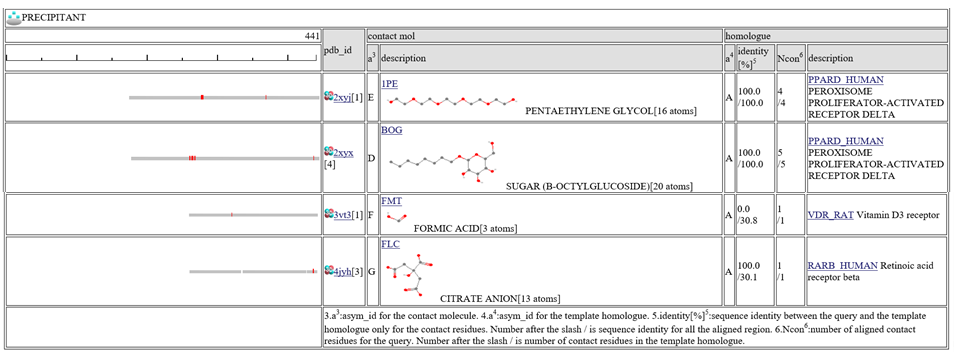
The homologous complexes are classified into seven classes: " HETERO", "
HETERO", " NUCLEOTIDE", "
NUCLEOTIDE", " COMPOUND", "
COMPOUND", " METAL", "
METAL", " OTHERPOLY", "
OTHERPOLY", " HOMO",
HOMO",  PRECIPITANT"".
PRECIPITANT"".
Aligned regions are shown in gray bars![]() , and residue contacting the other molecule are shown in small red boxes
, and residue contacting the other molecule are shown in small red boxes![]() .
.
If you click  3D icon, 3D structure will be displayed. Details of the 3D model viewer is described in another help page.
3D icon, 3D structure will be displayed. Details of the 3D model viewer is described in another help page.
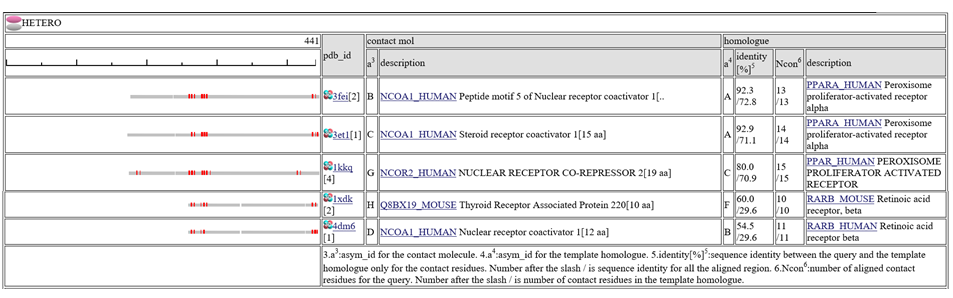
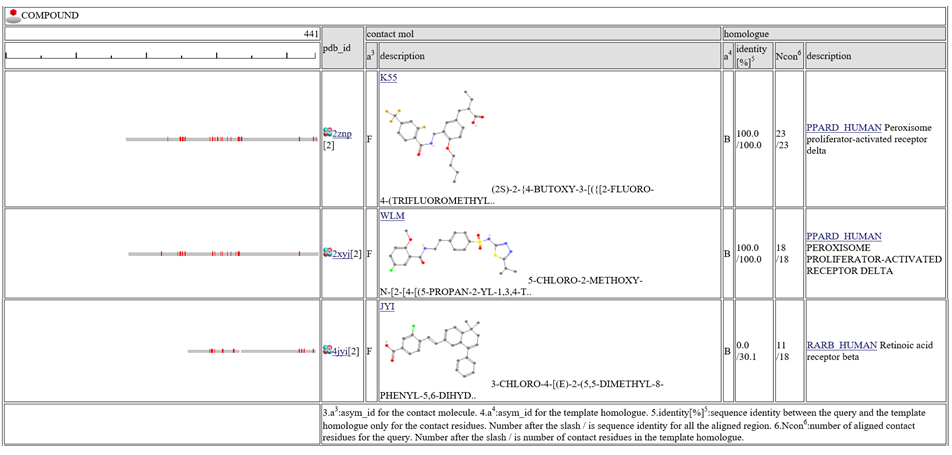
The column "contact_mol" in the middle of the table shows contacting molecules with query's homologues. The column "homologue" shows a homologous protein with the query. In the column "identity[%]", two sequence identities are shown, such as "92.3/72.3". The first number is the sequence identity of the contact site, and the second number is the identiy of all the aligned residues. The identity of the contact sites is a good measure for the qualit of the model, especially for the small chemical compound. In the column "Ncon", two number of contacting residues are shown, such as "13/13". The first number is the number of contact in the aligned regions, the second number is the number of contact sites in all the template sites. If the first number is much smaller than the second, the model can be built by a quite small part of the template complex 3D structures.
 "Site Table" view
"Site Table" view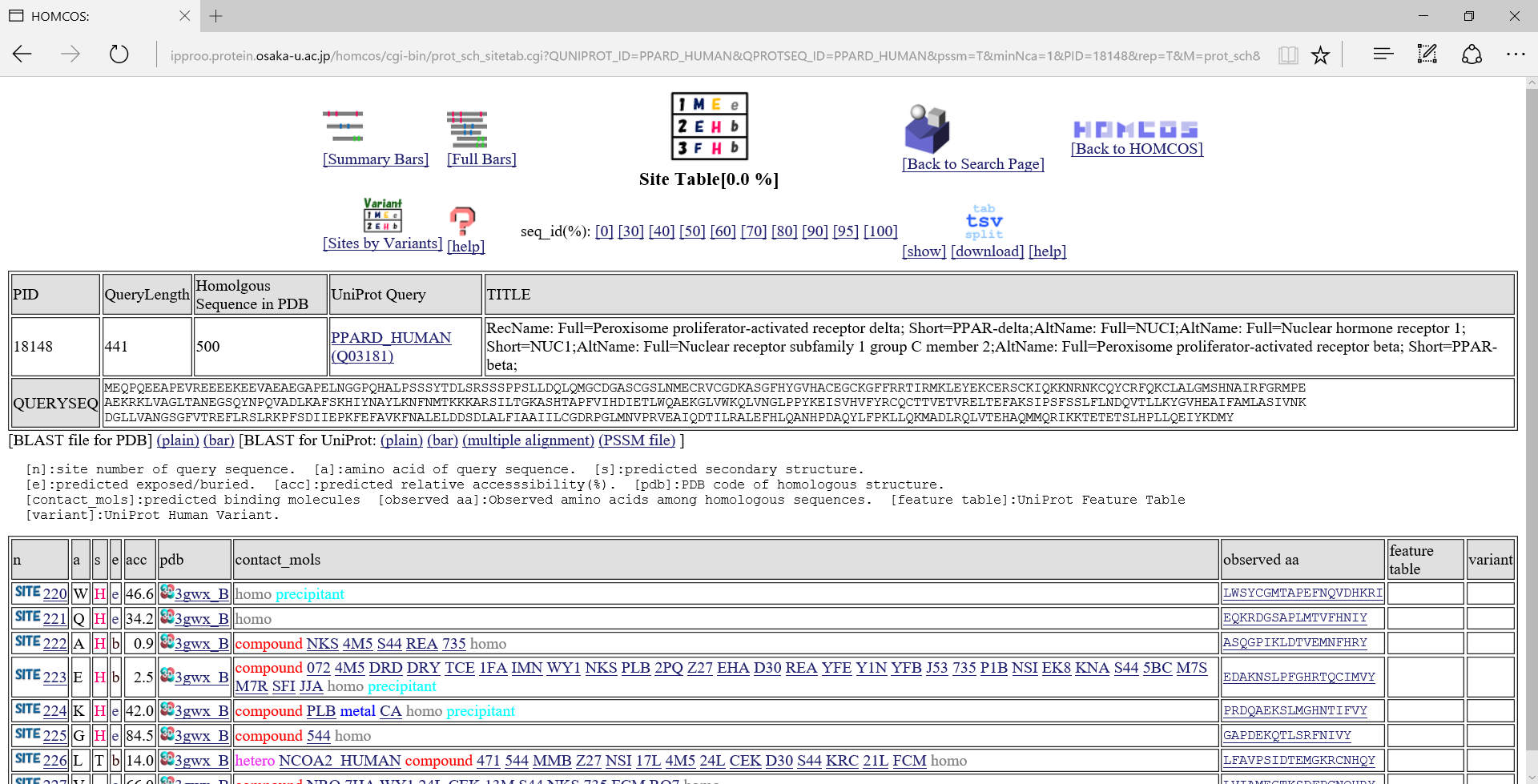
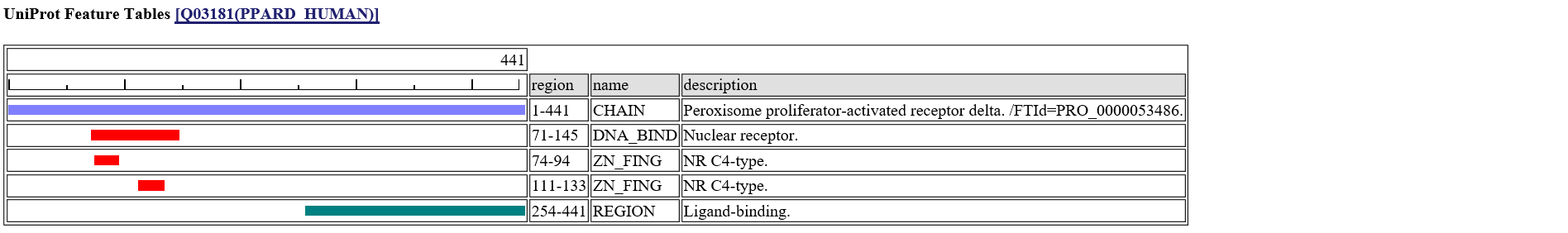
[n]:site number of query sequence. [a]:amino acid of query sequence. [s]:predicted secondary structure from homologous structure."H":alpha-helix, "E":beta-strand, "T":turn. [e]:predicted solvent accessible state. "e":exposed, "b":buried [acc]:predicted relative accesssibility[%]. [pdb]:PDB code of homologous structure. [contact_mols]:binding other molecules [observed aa]:Observed amino acids among homologous sequences. [feature table]:UniProt Feature Table [variant]:UniProt Human Variant.If you click the icon
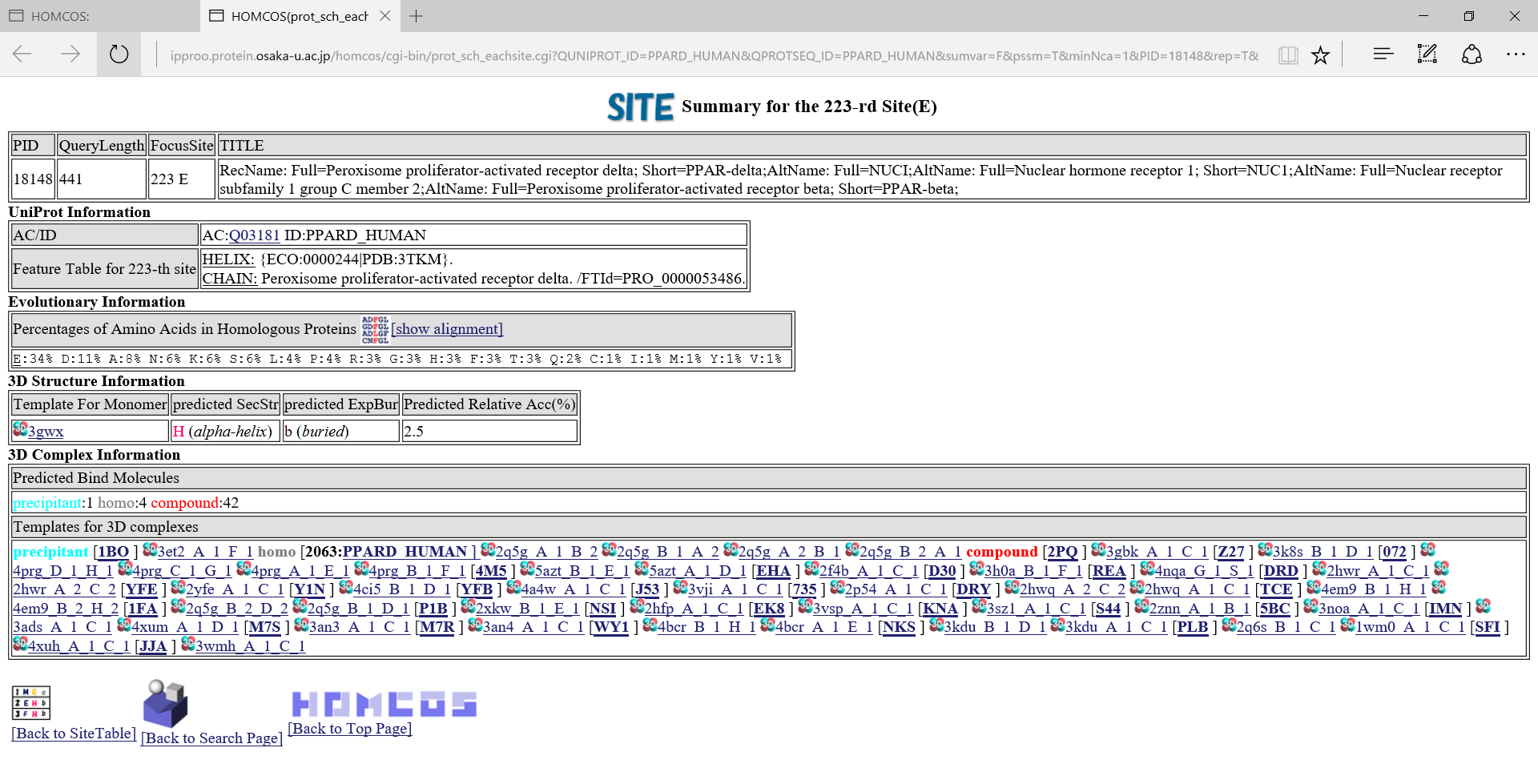
 in the left column, the summary page of the specific site appears.A following page is the summary of the 223-rd site of PPARD_HUMAN.
in the left column, the summary page of the specific site appears.A following page is the summary of the 223-rd site of PPARD_HUMAN.
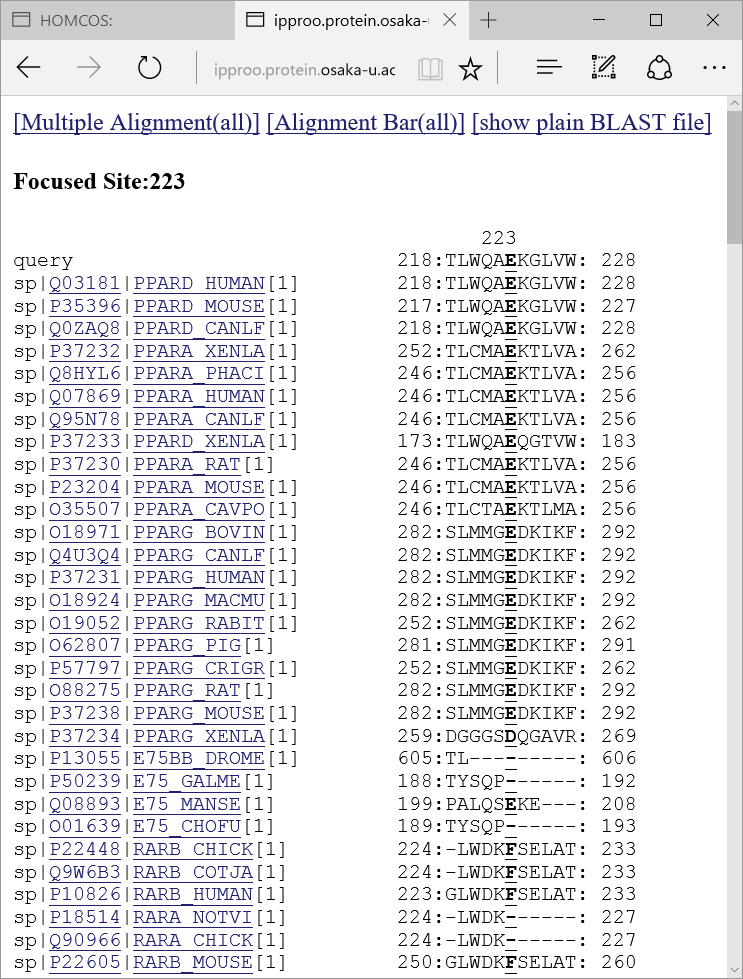
 , a following multiple alignment is shown about only the neighbor sites around the focused sites.
, a following multiple alignment is shown about only the neighbor sites around the focused sites.
If you click the icon  in "Templates for 3D complexes"、a following 3D model page is shown for the 3D complex model where the focused site contacts to other molecule.
in "Templates for 3D complexes"、a following 3D model page is shown for the 3D complex model where the focused site contacts to other molecule.
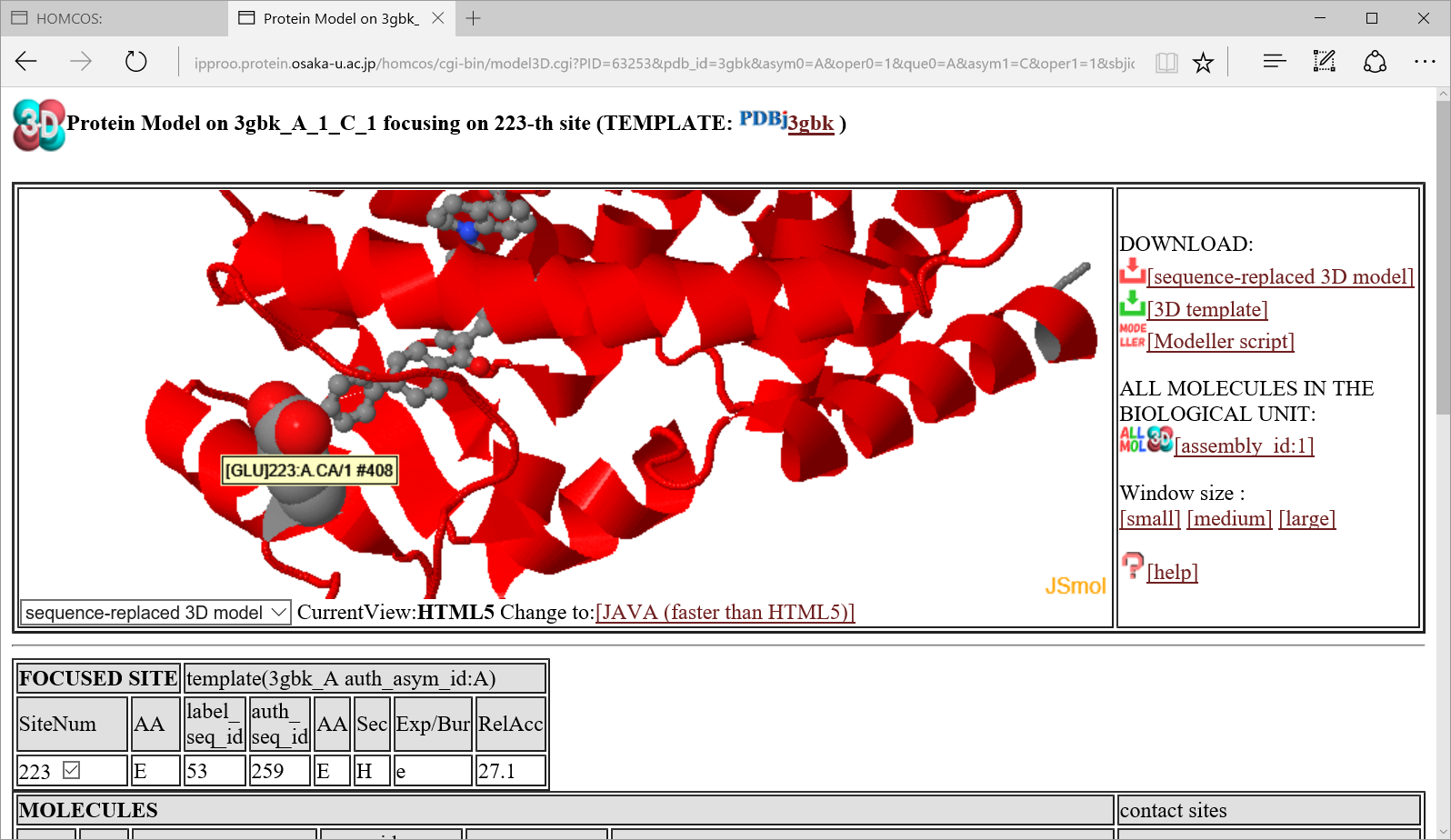
 how to use 3D model viewer in another help page.
how to use 3D model viewer in another help page.