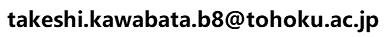- Overview of the search
- Input a query protein sequence
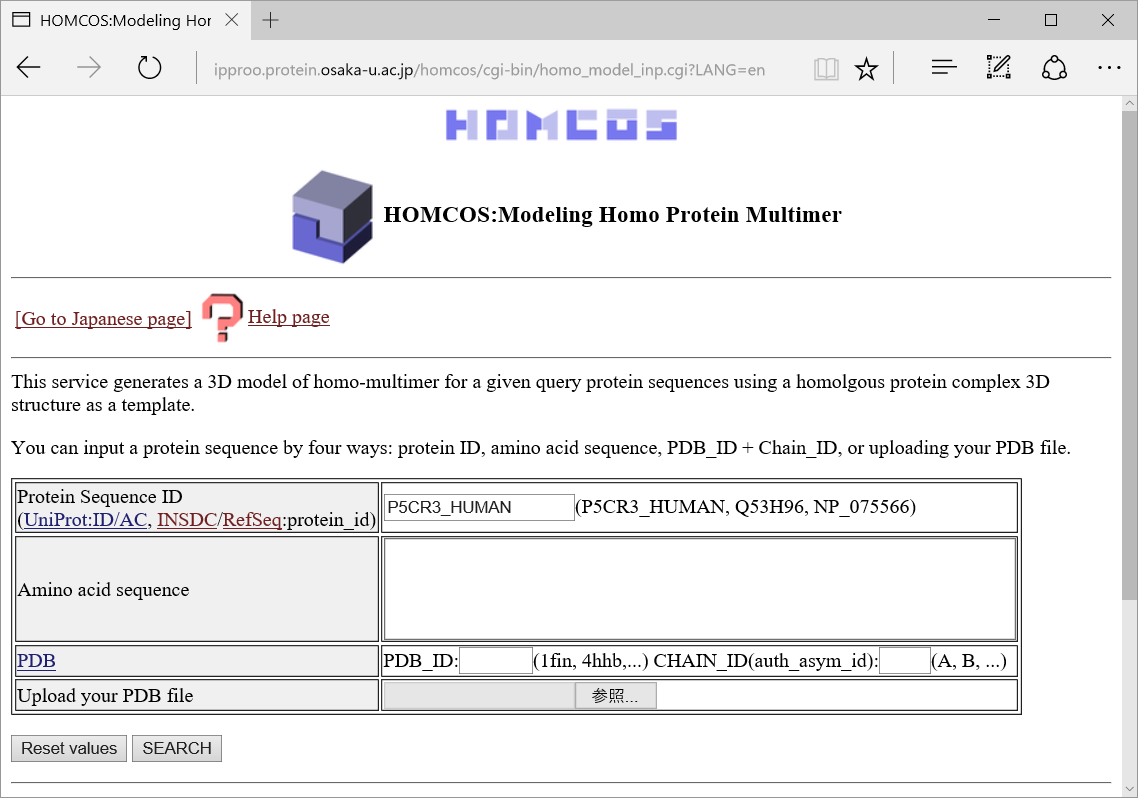 You can input a query protein sequence by following various methods.
You can input a query protein sequence by following various methods.
- ID for several protein sequnece databases
UniProt ID and AC, INSDC protein_id, RefSeq protein_id are available.
- amino acid sequence (one letter)
For example, "GIVEQCCASVCSLYQLENYCN".
- PDB_ID + CHAIN_ID
For example, PDB_ID="2ins" CHAIN_ID="A".
- Uploading your PDB file
Notes for input the query sequence
- HOMCOS has no string search services; protein names, gene names and keywords cannot be used as the query. If you know only names and keywords of the protein of your interest, please use other servers, such as UniProt, for string searches to get protein_id or amino acid sequences, before accessing HOMCOS.
- We recommended to use UniProt ID or AC for Human/Mouse/Rat queries. We can quickly show the search results of these sequences, because we precalculate BLAST searches for UniProts for these sequences.
- If 3D structure is assigned as the query, template-based docking is available using the query structure.
- Search results
This service is for modeling the complex 3D structures of homo multimers from one query protein. The server performs a seqnece homology searche (blastp;Altschul et al., 1997) with the given query protein sequence for all of the unitmol sequneces in the PDB, to make a list of the homologous proteins. The server then checks if a 3D multimeric structure of the homologues exists, in which subunit proteins are homologous to the query protein. If the homologous multimers are found, then they can be used as the template for complex modeling.
- Input a query protein sequence
 You can input a query protein sequence by following various methods.
You can input a query protein sequence by following various methods.
- ID for several protein sequnece databases
UniProt ID and AC, INSDC protein_id, RefSeq protein_id are available.
- amino acid sequence (one letter)
For example, "GIVEQCCASVCSLYQLENYCN".
- PDB_ID + CHAIN_ID
For example, PDB_ID="2ins" CHAIN_ID="A".
- Uploading your PDB file
Notes for input the query sequence
- HOMCOS has no string search services; protein names, gene names and keywords cannot be used as the query. If you know only names and keywords of the protein of your interest, please use other servers, such as UniProt, for string searches to get protein_id or amino acid sequences, before accessing HOMCOS.
- We recommended to use UniProt ID or AC for Human/Mouse/Rat queries. We can quickly show the search results of these sequences, because we precalculate BLAST searches for UniProts for these sequences.
- If 3D structure is assigned as the query, template-based docking is available using the query structure.
We will show a search result page of the homo modeling service for the query P5CR3_HUMAN.
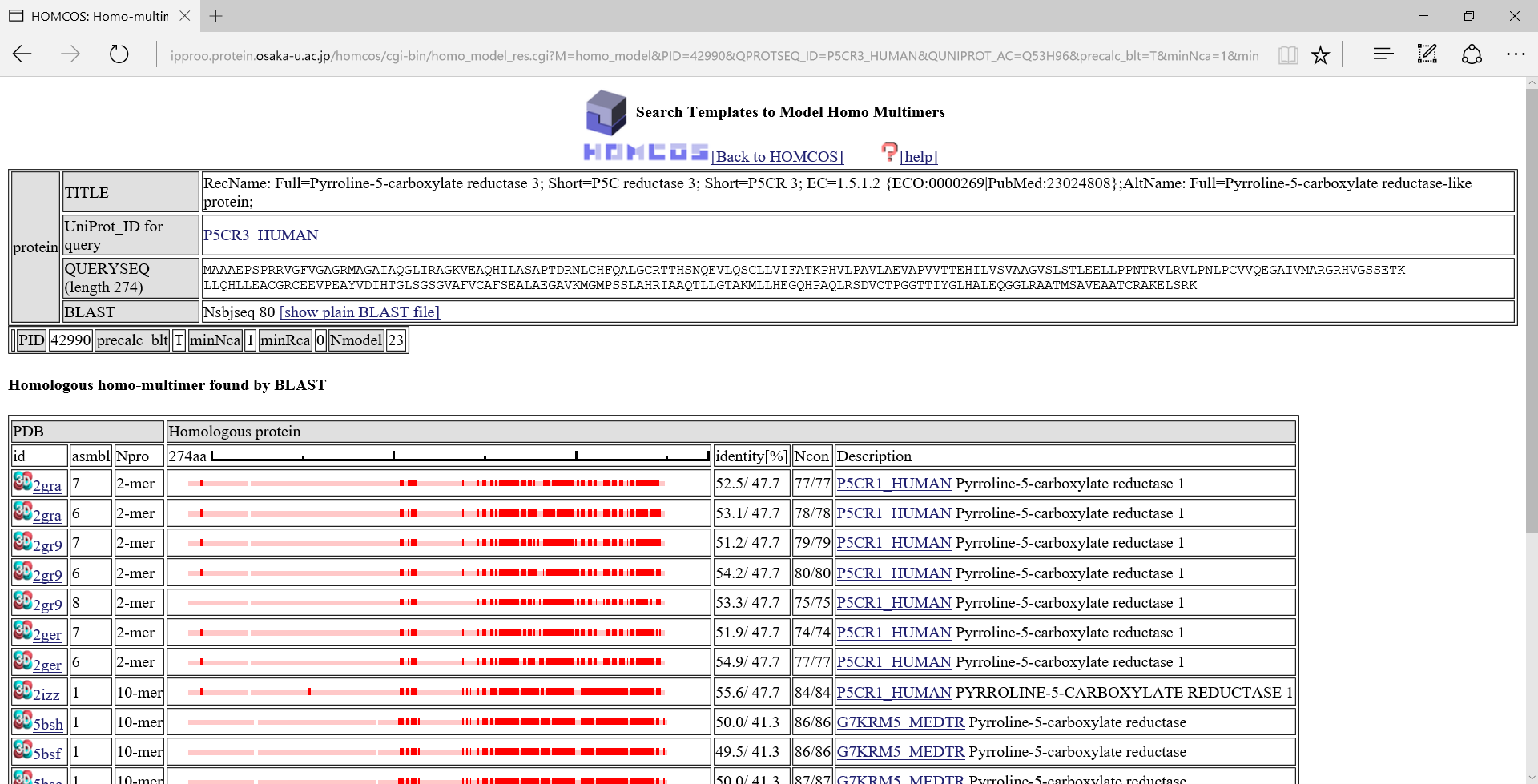 This page summarizes a results of the homology searche (blastp) for the query protein. Pale red bars
This page summarizes a results of the homology searche (blastp) for the query protein. Pale red bars  in the left are aligned regions for the query protein. Deep red boxes
in the left are aligned regions for the query protein. Deep red boxes  show contacting residues.
show contacting residues.
If one of the 3D icons  in the left column is clicked, a following 3D model viewer window will be displayed.
in the left column is clicked, a following 3D model viewer window will be displayed.
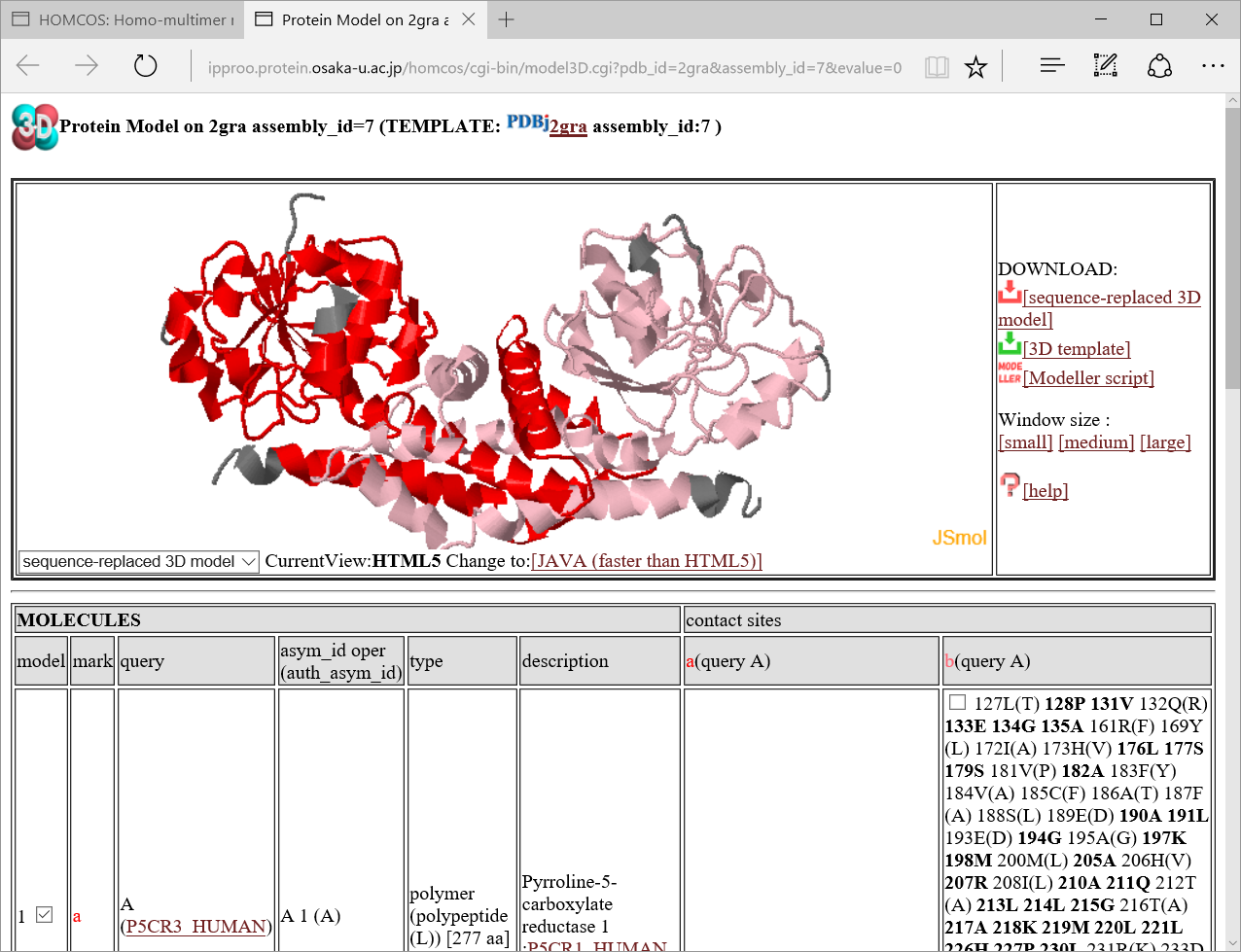 We will explain
We will explain  how to use 3D model viewer in another help page.
how to use 3D model viewer in another help page.


 Homo Protein Multimer
Homo Protein Multimer


 Homo Protein Multimer
Homo Protein Multimer
 Homo Protein Multimer
Model 3D structure of homo-multimer for one given sequence using homology-modeling technique.
Homo Protein Multimer
Model 3D structure of homo-multimer for one given sequence using homology-modeling technique.




 in the left column is clicked, a following 3D model viewer window will be displayed.
in the left column is clicked, a following 3D model viewer window will be displayed.

 how to use 3D model viewer in another help page.
how to use 3D model viewer in another help page.