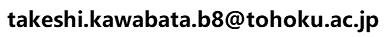[Go to Japanese page]
Many mutations are observed in cancer cells, it is important to extract functionally significant sites among them. This page explains, how to identify potential cancer-associated sites by "3D cluster" sites (dense mutations site in 3D space) with the help of HOMCOS server and PyMOL script py3Dcluster.py. The algorithm for detecting 3D cluster is based on Gao et al, 2017.
Purpose
Identify potential cancer associated sites as mutation sites clustered in 3D space with statistically significance (3D cluster), and the neighboring surface sites around the 3D cluster.
Procedures
[1]:Search mutations from "cBioPortal" for your query protein, and save them as the tsv file.
[2]:Extract "ProteinChange" column by a spreadsheet application.
[3]:Search and model a 3D model for the query protein by HOMCOS server, save it as a PDB file
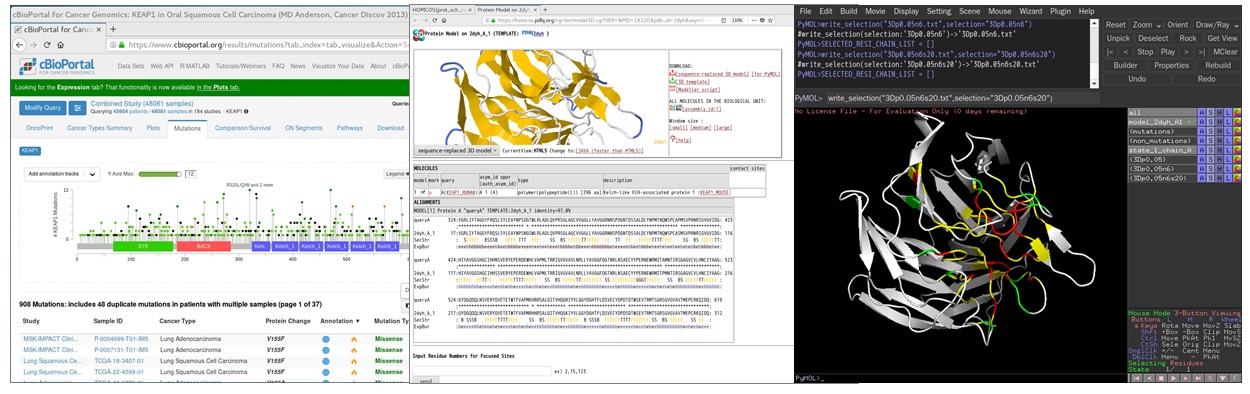
[4]:Analyze the 3D model using PyMOL with the help of the script "py3Dcluster.py"
This work was supported by the Project of Osaka University Institute for Datability Science.
LastModfied:2023/01/31
Questions and comments: