|
|
| PID | QueryLength | FocusSite | TITLE |
| 31440 | 4405 | 3326 N | RecName: Full=Replicase polyprotein 1a; Short=pp1a;AltName: Full=ORF1a polyprotein;Contains: RecName: Full=Host translation inhibitor nsp1; AltName: Full=Leader protein; AltName: Full=Non-structural protein 1; Short=nsp1;Contains: RecName: Full=Non-structural protein 2; Short=nsp2; AltName: Full=p65 homolog;Contains: RecName: Full=Papain-like protease nsp3 ; EC=3.4.19.12 ; EC=3.4.22.-; AltName: Full=Non-structural protein 3; Short=nsp3; AltName: Full=PL2-PRO; AltName: Full=Papain-like proteinase; Short=PL-PRO;Contains: RecName: Full=Non-structural protein 4; Short=nsp4;Contains: RecName: Full=3C-like proteinase nsp5; Short=3CL-PRO; Short=3CLp; EC=3.4.22.69; AltName: Full=Main protease; Short=Mpro ; AltName: Full=Non-structural protein 5; Short=nsp5; AltName: Full=SARS coronavirus main proteinase;Contains: RecName: Full=Non-structural protein 6; Short=nsp6;Contains: RecName: Full=Non-structural protein 7; Short=nsp7;Contains: RecName: Full=Non-structural protein 8; Short=nsp8;Contains: RecName: Full=RNA-capping enzyme subunit nsp9; AltName: Full=Non-structural protein 9; Short=nsp9; EC=2.7.7.50 ;Contains: RecName: Full=Non-structural protein 10; Short=nsp10; AltName: Full=Growth factor-like peptide; Short=GFL;Contains: RecName: Full=Non-structural protein 11; Short=nsp11; |
| AC/ID | AC:P0DTC1 ID:R1A_SARS2 |
| Feature Table for 3326-th site |
HELIX: CHAIN: /note="3C-like proteinase nsp5" /id="PRO_0000449639" DOMAIN: /note="Peptidase C30" TOPO_DOM: /note="Cytoplasmic" CHAIN: /note="Replicase polyprotein 1a" /id="PRO_0000449634" |
Percentages of Amino Acids in Homologous Proteins
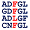 [show alignment] [show alignment]
|
N:48% L:23% S:15% A:1% R:1% D:1% Q:1% E:1% G:1% I:1% K:1% F:1% P:1% T:1% Y:1% V:1% |
| Template For Monomer | predicted SecStr | predicted ExpBur | Predicted Relative Acc(%) |
 6xbi 6xbi |
G (3/10-helix) | e (exposed) | 31.5 |
| Predicted Bind Molecules |
| compound:4 metal:4 precipitant:17 |
| Templates for 3D complexes |
compound [K3S ]  5rfb_A_1_E_1 5rfb_A_1_E_1  5rfb_A_2_E_2 [A82 ] 5rfb_A_2_E_2 [A82 ]  6yvf_A_1_B_1 6yvf_A_1_B_1  6yvf_A_2_B_2
metal [NA ] 6yvf_A_2_B_2
metal [NA ]  6xbh_A_1_D_1 6xbh_A_1_D_1  6xbh_A_2_D_2 6xbh_A_2_D_2  7lyi_A_1_C_1 7lyi_A_1_C_1  7lyi_A_2_C_2
precipitant [GOL ] 7lyi_A_2_C_2
precipitant [GOL ]  6xa4_A_1_C_1 6xa4_A_1_C_1  6xa4_A_2_C_2 6xa4_A_2_C_2  6xbg_B_1_E_1 6xbg_B_1_E_1  6xbh_A_1_C_1 6xbh_A_1_C_1  6xbh_A_2_C_2 6xbh_A_2_C_2  6xfn_A_1_C_1 6xfn_A_1_C_1  6xfn_A_2_C_2 6xfn_A_2_C_2  7lyi_A_1_B_1 7lyi_A_1_B_1  7lyi_A_2_B_2 7lyi_A_2_B_2  7rn1_A_1_B_1 7rn1_A_1_B_1  7rn1_A_2_B_2 7rn1_A_2_B_2  7t8m_B_1_N_1 [DMS ] 7t8m_B_1_N_1 [DMS ]  7grs_B_1_DA_1 [PEG ] 7grs_B_1_DA_1 [PEG ]  7tob_A_1_C_1 7tob_A_1_C_1  7tob_A_2_C_2 [EDO ] 7tob_A_2_C_2 [EDO ]  8ute_A_1_J_1 8ute_A_1_J_1  8ute_A_2_J_2 8ute_A_2_J_2
|
 [Back to SiteTable] |
 [Back to Search Page] |
 [Back to Top Page] |