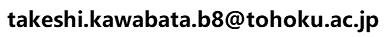- Overview of the search
 This service is for modeling the complex 3D structure of one protein and one small chemical compound. A user can input a query protein as its amino acid sequence or its 3D structures, as with the other services require. In addition, a query chemical structure is input by various methods. For the 3D modeling, the uploaded chemical structure file should not be 2D, but should have a 3D conformation with sufficiently quality (proper bond lengthes and bond angles)for the initial conformation. The server performs a sequence homology search ( blastp;Altschul et al., 1997 ) with one given query protein sequence for all of the unitmol sequences in the PDB, to make a list of the homologous proteins. The server also performs a chemical similarity search with the given query chemical compound for all of the chemical compounds appearing in the PDB, by the dkcombu program (Kawabata et al., 2013). The conformation of the query protein is modelled in the same way to the other services: simple sequence-replacing model or input query 3D structure. The conformation of the query compound is modelled by our flexible superposition program fkcombu(Kawabata and Nakamura, 2014). The program fkcombu aligns the query target compound based on the atomic correspondences with the template compound, by changing the conformation of the target. The atomic correspondence are calculated by the maximum commonn substructure (MCS) algorithm.
This service is for modeling the complex 3D structure of one protein and one small chemical compound. A user can input a query protein as its amino acid sequence or its 3D structures, as with the other services require. In addition, a query chemical structure is input by various methods. For the 3D modeling, the uploaded chemical structure file should not be 2D, but should have a 3D conformation with sufficiently quality (proper bond lengthes and bond angles)for the initial conformation. The server performs a sequence homology search ( blastp;Altschul et al., 1997 ) with one given query protein sequence for all of the unitmol sequences in the PDB, to make a list of the homologous proteins. The server also performs a chemical similarity search with the given query chemical compound for all of the chemical compounds appearing in the PDB, by the dkcombu program (Kawabata et al., 2013). The conformation of the query protein is modelled in the same way to the other services: simple sequence-replacing model or input query 3D structure. The conformation of the query compound is modelled by our flexible superposition program fkcombu(Kawabata and Nakamura, 2014). The program fkcombu aligns the query target compound based on the atomic correspondences with the template compound, by changing the conformation of the target. The atomic correspondence are calculated by the maximum commonn substructure (MCS) algorithm.
- Input one query protein sequence and one chemical compound
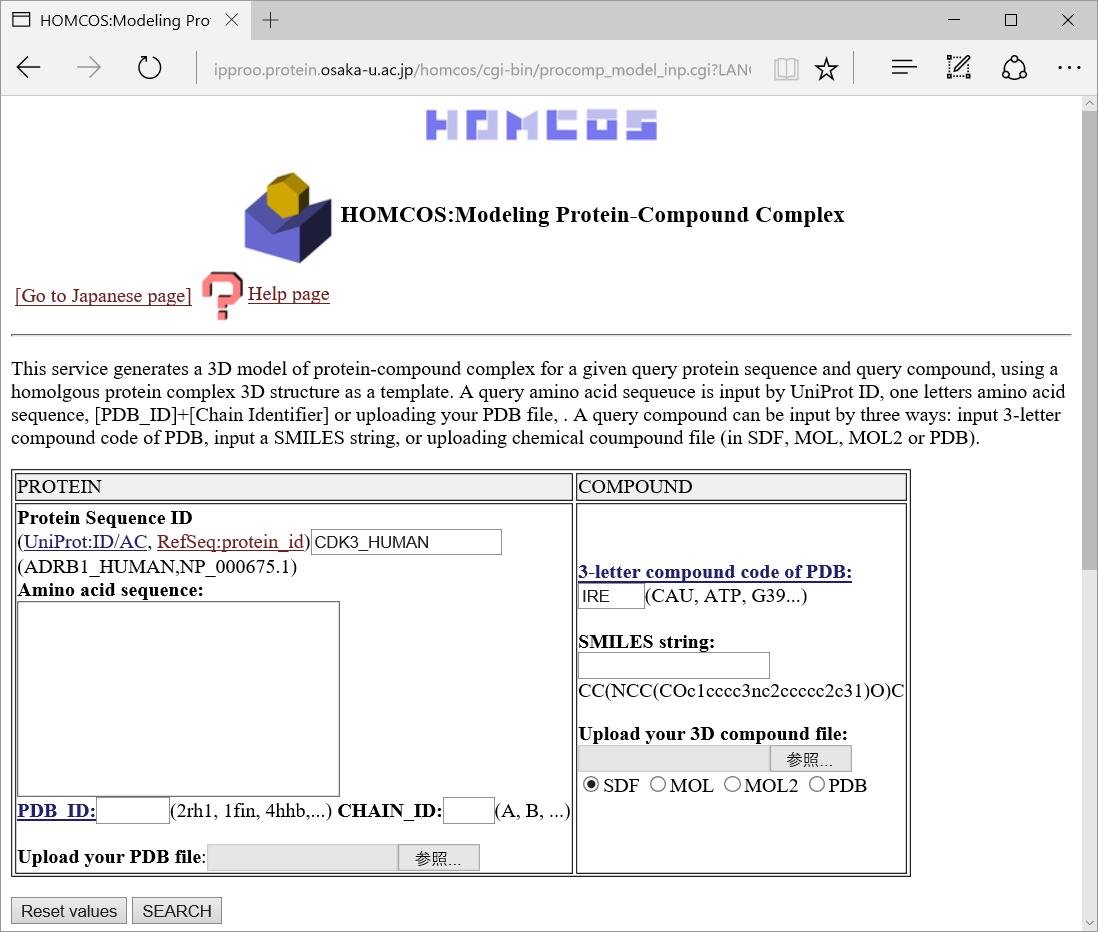 Input one query protein sequence
Input one query protein sequence
You can input a query protein sequence by following various methods.
- ID for several protein sequnece databases
UniProt ID and AC, INSDC protein_id, RefSeq protein_id are available.
- amino acid sequence (one letter)
For example, "GIVEQCCASVCSLYQLENYCN".
- PDB_ID + CHAIN_ID
For example, PDB_ID="2ins" CHAIN_ID="A".
- Uploading your PDB file
Notes for input the query sequence
- HOMCOS has no string search services; protein names, gene names and keywords cannot be used as the query. If you know only names and keywords of the protein of your interest, please use other servers, such as UniProt, for string searches to get protein_id or amino acid sequences, before accessing HOMCOS.
- We recommended to use UniProt ID or AC for Human/Mouse/Rat queries. We can quickly show the search results of these sequences, because we precalculate BLAST searches for UniProts for these sequences.
- If 3D structure is assigned as the query, template-based docking is available using the query structure.
Input one query compound
You can input a query compound structure by following various methods.
- 3-letter compound code of PDB
Each chemical structure stored in PDB has its own unique 3-letter compound code. For example, the code for "ADENOSINE-5'-TRIPHOSPHATE", "BETA-D-GALACTOSE" and "Gefitinib" is "ATP", "GAL" and "IRE", respectively. You can search the 3-letter code for the molecular name using several serveres, such as Ligand Expo and PDBj Chemie search.
- SMILES string
A string representation of SMILES is useful for copy and pasting chemical structures. Many chemical databases provides SMILES string representaions. The SMILES string of glycine and phenylalanine is "NCC(=O)O" and "OC(C(Cc1ccccc1)N)=O", respectively. Details of the SMILES string is described in wiki.
- Uploading a chemical compound file
Users can upload their own chemical structure file. Four formats, SDF, MOL, MOL2 and PDB are available.
CAUTION: an uploading compound file should have a 3D conformation with proper bond lengthes and angles. A planar 2D structure is not good for 3D modeling. If a SMILES string is used as the query, the server makes its 2D structure and use it for the 3D modeling.
- Search results
We will show a search result page of the protein-compound modeling service for the queries CDK3_HUMAN and IRE.
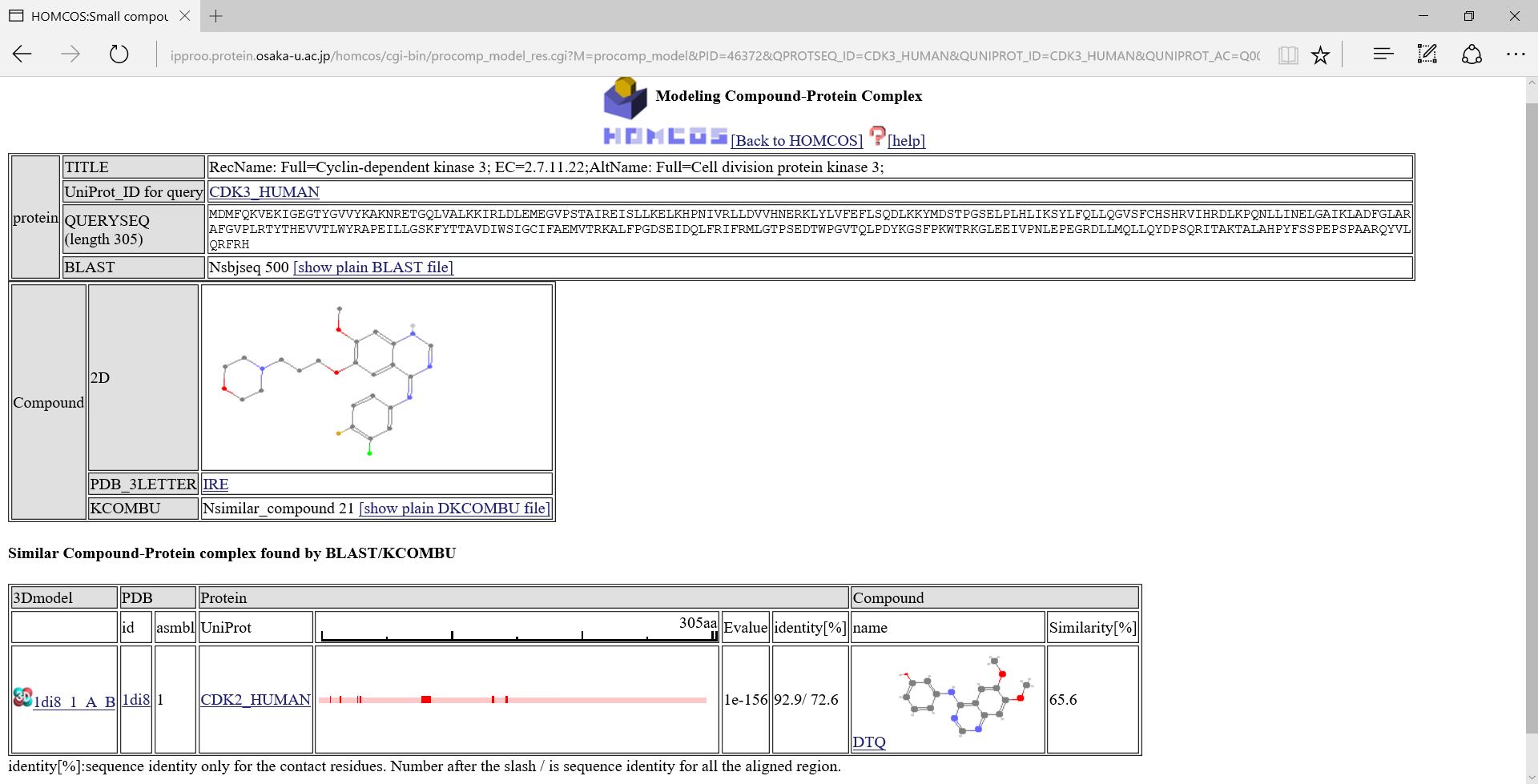
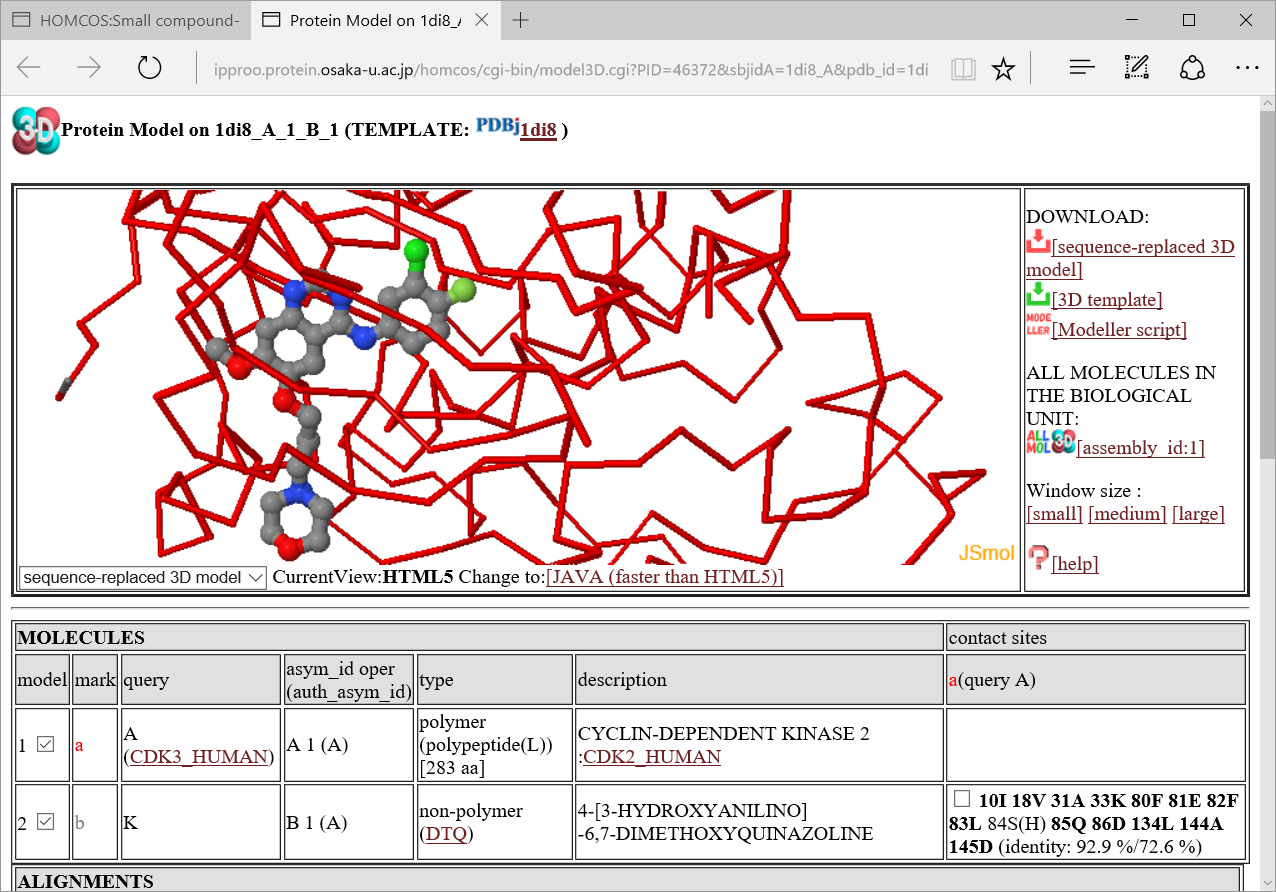
If one of the 3D icons  in the left column is clicked, a following 3D model viewer window will be displayed.
in the left column is clicked, a following 3D model viewer window will be displayed.
If you chose the option "template 3D structure" of the menu in the left, you can see the template chemical structure(DTQ).
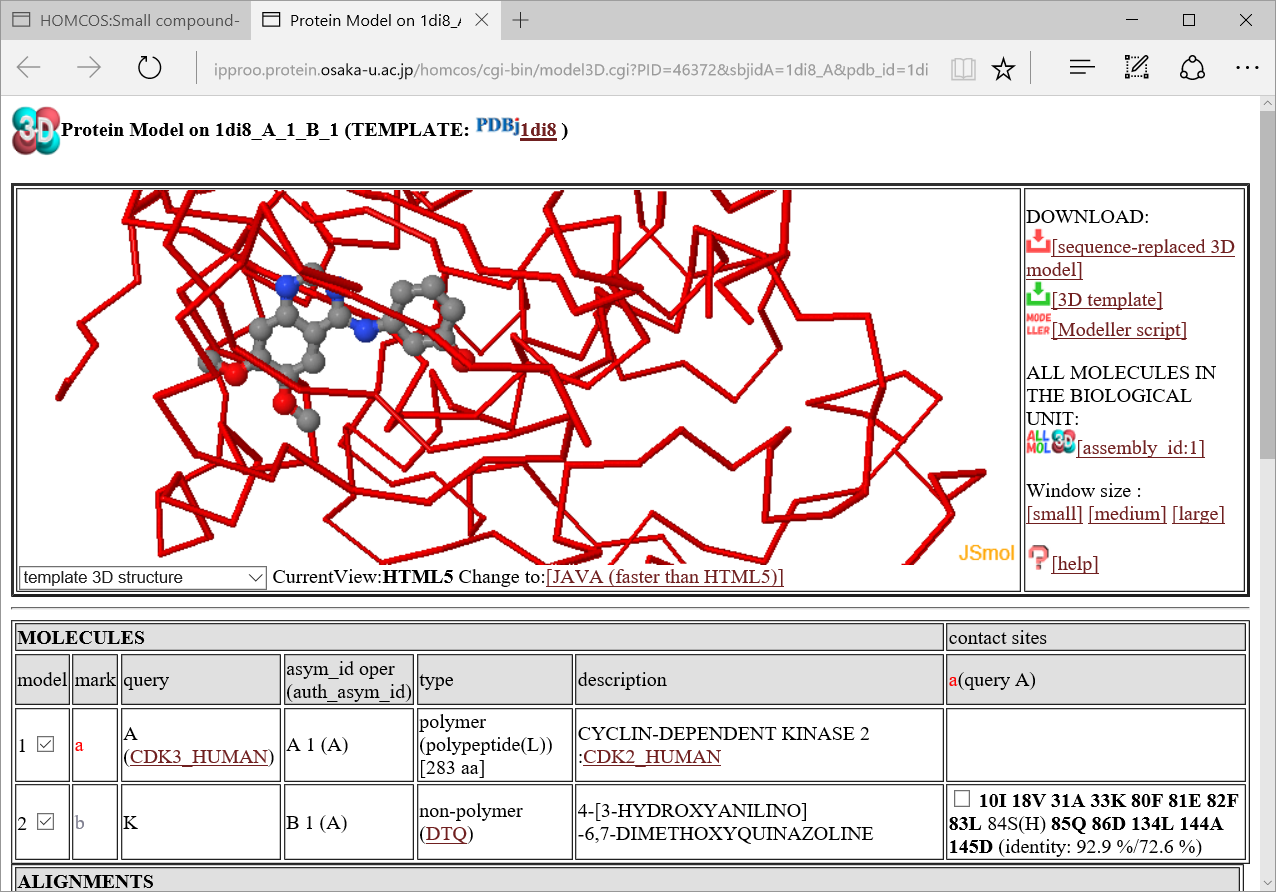 We will explain
We will explain  how to use 3D model viewer in another help page.
how to use 3D model viewer in another help page.


 Protein-Compound Complex
Protein-Compound Complex


 Protein-Compound Complex
Protein-Compound Complex
 Protein-Compound Complex
Model 3D structure of protein-compound complex for one given sequence and one given chemical structure using homology-modeling technique.
Protein-Compound Complex
Model 3D structure of protein-compound complex for one given sequence and one given chemical structure using homology-modeling technique.



 in the left column is clicked, a following 3D model viewer window will be displayed.
in the left column is clicked, a following 3D model viewer window will be displayed.


 how to use 3D model viewer in another help page.
how to use 3D model viewer in another help page.