|
|||
 [Back to HOMCOS] |
[DKCOMBU] |
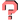 [help] |
 [show] [download] |
| threshold chemical similarty (tanimoto coefficient) |
[0.5] [0.6] [0.7] [0.8] [0.9] [1.0] | max number of compounds | [1] [5] [10] [20] [30] [40] [50] |
|
|||
 [Back to HOMCOS] |
[DKCOMBU] |
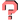 [help] |
 [show] [download] |
| threshold chemical similarty (tanimoto coefficient) |
[0.5] [0.6] [0.7] [0.8] [0.9] [1.0] | max number of compounds | [1] [5] [10] [20] [30] [40] [50] |
| compound | similarity | compound | similarity | compound | similarity | compound | similarity | compound | similarity |
| query [2D] [3D] |
*** | 0TX | 1.000 | CLQ | 1.000 | QUN | 0.786 | QUM | 0.733 |
| 1O8 | 0.630 | CQA | 0.621 | E2C | 0.621 | 29V | 0.607 | 2VR | 0.607 |
| 2VP | 0.607 | 2VZ | 0.607 | 3RB | 0.607 | R4M | 0.607 | A8N | 0.586 |
| RTX | 0.586 | RNP | 0.577 | SNP | 0.577 | 2W0 | 0.552 | C4Q | 0.552 |
| HLH | 0.538 | W45 | 0.536 | 1PQ | 0.519 | HQO | 0.519 | HQ | 0.519 |
| JWW | 0.519 | SP9 | 0.519 | SP8 | 0.519 | 30M | 0.517 | B9O | 0.517 |
| B9R | 0.517 | CAU | 0.517 | 4XG | 0.500 | AUX | 0.500 | BFA | 0.500 |
| IHQ | 0.500 | KWK | 0.500 | NNQ | 0.500 | WW9 | 0.500 | TNH | 0.486 |
| 0NI | 0.485 |
| Nrank | Protein | Nresidue | pdb_id+asym_id |
| 1 | Q9U7N7_PLAFA Falcilysin | 1076 |
[CLQ]: 7di7_A_D 7di7_A_D
|
| 2 | BRD4_HUMAN Bromodomain-containing protein 4 | 127 |
[30M]: 4qb3_A_B 4qb3_A_B
|
| 3 | TNNC1_HUMAN:TNNI3_HUMAN Troponin C, slow skeletal and cardiac muscles,Troponin I, cardiac muscle | 125 |
[WW9]: 7uh9_A_C 7uh9_A_C
|
| 4 | CD44_MOUSE CD44 antigen | 150 |
[4XG]: 5bzp_A_B 5bzp_A_B
|
| 5 | A5K867_PLAVS Phosphoethanolamine N-methyltransferase, putative | 265 |
[CQA]: 4mwz_B_H 4mwz_B_H
|
| 6 | ADRB2_HUMAN Beta-2 adrenergic receptor | 286 |
[CAU]: 4gbr_A_C 4gbr_A_C
|
| 7 | TRFL_BOVIN Lactotransferrin | 341 |
[RNP]: 3mjn_A_J 3mjn_A_J
|
| 8 | A0A077KEB8_9ACTN Cytochrome P-450 | 392 |
[B9R]: 6j83_A_C 6j83_A_C
|
| 9 | GUX1_TRIRE EXOGLUCANASE 1 | 434 |
[SNP]: 1dy4_A_D 1dy4_A_D
|
| 10 | Q09431_PHACH EXOGLUCANASE I | 431 |
[RNP]: 1h46_A_C 1h46_A_C
|
| 11 | A9WEV4_CHLAA Polysulphide reductase NrfD | 449 |
[HQO]: 8x2j_C_S 8x2j_C_S
|
| 12 | A9WEV3_CHLAA Fe-S-cluster-containing hydrogenase components 1-like protein | 951 |
[HQO]: 8x2j_B_S 8x2j_B_S
|
| 13 | FRDD_ECOLI FUMARATE REDUCTASE 13 KDA HYDROPHOBIC PROTEIN | 119 |
[HQO]: 1kf6_D_R
[HQO]: 1kf6_D_R
[HQO]: 1kf6_H_Z 1kf6_H_Z
|
| 14 | FRDC_ECOLI FUMARATE REDUCTASE 15 KDA HYDROPHOBIC PROTEIN | 130 |
[HQO]: 1kf6_C_R
[HQO]: 1kf6_C_R
[HQO]: 1kf6_G_Z 1kf6_G_Z
|
| 15 | Q72EF4_DESVH NAPC/NIRT CYTOCHROME C FAMILY PROTEIN | 144 |
[HQO]: 2vr0_C_Y
[HQO]: 2vr0_C_Y
[HQO]: 2vr0_F_RA 2vr0_F_RA
|
| 16 | FRDB_ECOLI FUMARATE REDUCTASE IRON-SULFUR PROTEIN | 243 |
[HQO]: 1kf6_B_R
[HQO]: 1kf6_B_R
[HQO]: 1kf6_F_Z 1kf6_F_Z
|
| 17 | PIM1_HUMAN Serine/threonine-protein kinase pim-1 | 273 |
[E2C]: 6l11_A_B
[RTX]: 6l11_A_B
[RTX]: 6kzi_A_B 6kzi_A_B
|
| 18 | ADRB1_MELGA BETA-1 ADRENERGIC RECEPTOR | 297 |
[CAU]: 2ycw_A_C
[CAU]: 2ycw_A_C
[CAU]: 2ycw_B_G 2ycw_B_G
|
| 19 | A7YT55_DANRE Hdac6 protein | 355 |
[W45]: 6mr5_A_F
[W45]: 6mr5_A_F
[W45]: 6mr5_B_K 6mr5_B_K
|
| 20 | PYRD_ECOLI Dihydroorotate dehydrogenase (quinone) | 335 |
[HQO]: 7t5k_A_D
[HQO]: 7t5k_A_D
[HQO]: 7t5k_B_F 7t5k_B_F
|
| 21 | F5L3B8_9BACI FAD-dependent pyridine nucleotide-disulfide oxidoreductase | 394 |
[HQO]: 6bdo_A_F
[HQO]: 6bdo_A_F
[HQO]: 6bdo_D_J 6bdo_D_J
|
| 22 | A0A085SSI3_VIBCL Na(+)-translocating NADH-quinone reductase subunit B | 412 |
[HQO]: 8a1y_B_N
[0NI]: 8a1y_B_N
[0NI]: 7xk6_B_I 7xk6_B_I
|
| 23 | A0A1M4Y7D5_STRHI Nocardicin N-oxygenase | 395 |
[B9O]: 6j88_A_D
[B9O]: 6j88_A_D
[B9O]: 6j88_B_F 6j88_B_F
|
| 24 | A0A655PZA5_VIBCL Na(+)-translocating NADH-quinone reductase subunit A | 448 |
[HQO]: 8a1y_A_N
[0NI]: 8a1y_A_N
[0NI]: 7xk6_A_I 7xk6_A_I
|
| 25 | D9IEF7_BPT4 Endolysin,Beta-1 adrenergic receptor chimera | 455 |
[CAU]: 7bvq_A_E
[CAU]: 7bvq_A_E
[CAU]: 7bvq_B_S 7bvq_B_S
|
| 26 | ENLYS_BPT4:ADRB2_HUMAN Endolysin,Beta-2 adrenergic receptor | 391 |
[CAU]: 5jqh_A_E
[CAU]: 5jqh_A_E
[CAU]: 5jqh_B_G 5jqh_B_G
|
| 27 | APPC_ECOLI Cytochrome bd-II ubiquinol oxidase subunit 1 | 503 |
[0NI]: 7ose_A_J
[0NI]: 7ose_A_J
[0NI]: 7ose_D_O 7ose_D_O
|
| 28 | B3Y963_GEOSE Nitric oxide reductase | 754 |
[HQO]: 3ayg_A_K
[HQO]: 3ayg_A_K
[HQO]: 3ayg_A_K 3ayg_A_K
|
| 29 | FDNI_ECOLI FORMATE DEHYDROGENASE, NITRATE-INDUCIBLE, CYTOCHROME B556(FDN) SUBUNIT | 216 |
[HQO]: 1kqg_C_O
[HQO]: 1kqg_C_O
[HQO]: 1kqg_C_O
[HQO]: 1kqg_C_O
[HQO]: 1kqg_C_O 1kqg_C_O
|
| 30 | MROUPO | 234 |
[SNP]: 5fuk_A_F
[SNP]: 5fuk_A_F
[SNP]: 5fuk_B_F
[SNP]: 5fuk_B_F
[SNP]: 5fuk_B_L 5fuk_B_L
|
| 31 | B1MFK2_MYCA9 Putative dioxygenase (1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase) | 267 |
[JWW]: 6rb3_A_D
[JWW]: 6rb3_A_D
[JWW]: 6rb3_B_E
[JWW]: 6rb3_B_E
[JWW]: 6rb3_C_F 6rb3_C_F
|
| 32 | FMS1_YEAST fms1 | 492 |
[SP8]: 3cns_A_D
[SP8]: 3cns_A_D
[SP8]: 3cns_B_F
[SP9]: 3cns_B_F
[SP9]: 3cnt_A_D 3cnt_A_D
|
| 33 | B5B0D4_BOVIN Major allergen beta-lactoglobulin | 162 |
[0TX]: 7er3_A_E
[0TX]: 7er3_A_E
[0TX]: 7er3_B_F
[0TX]: 7er3_B_F
[0TX]: 7er3_C_G
[0TX]: 7er3_C_G
[0TX]: 7er3_D_H 7er3_D_H
|
| 34 | Q9I4X0_PSEAE Transcriptional regulator MvfR | 204 |
[HLH]: 6q7u_A_B
[HLH]: 6q7u_A_B
[HLH]: 6q7u_A_B
[NNQ]: 6q7u_A_B
[NNQ]: 4jvd_A_B
[NNQ]: 4jvd_A_B
[NNQ]: 4jvd_A_B 4jvd_A_B
|
| 35 | Q6T755_PLAFA Phosphoethanolamine N-methyltransferase | 258 |
[CQA]: 4fgz_A_C
[CQA]: 4fgz_A_C
[CQA]: 4fgz_A_D
[CQA]: 4fgz_A_D
[CQA]: 4fgz_B_G
[CQA]: 4fgz_B_G
[CQA]: 4fgz_B_H 4fgz_B_H
|
| 36 | LDH1_PLAFD PROTEIN (L-LACTATE DEHYDROGENASE) | 305 |
[CLQ]: 1cet_A_B
[CLQ]: 1cet_A_B
[CLQ]: 1cet_A_B
[CLQ]: 1cet_A_B
[CLQ]: 1cet_A_B
[CLQ]: 1cet_A_B
[CLQ]: 1cet_A_B 1cet_A_B
|
| 37 | APO1_AGRAE Aromatic peroxygenase | 325 |
[SNP]: 6ekz_A_N
[SNP]: 6ekz_A_N
[SNP]: 6ekz_A_O
[SNP]: 6ekz_A_O
[SNP]: 6ekz_A_P
[SNP]: 6ekz_A_P
[SNP]: 6ekz_A_Q 6ekz_A_Q
|
| 38 | ACES_TORCA ACETYLCHOLINESTERASE | 528 |
[A8N]: 1ut6_A_D
[A8N]: 1ut6_A_D
[A8N]: 1ut6_A_D
[TNH]: 1ut6_A_D
[TNH]: 4x3c_A_D
[TNH]: 4x3c_A_D
[TNH]: 4x3c_A_D 4x3c_A_D
|
| 39 | A0A063X8D0_BACIU AA3-600 quinol oxidase subunit I | 607 |
[HQO]: 6koe_A_L
[HQO]: 6koe_A_L
[HQO]: 6koe_H_P
[IHQ]: 6koe_H_P
[IHQ]: 6koc_A_L
[IHQ]: 6koc_A_L
[IHQ]: 6koc_H_P 6koc_H_P
|
| 40 | HNMT_HUMAN Histamine N-Methyltransferase | 280 |
[QUN]: 1jqe_A_D
[CQA]: 1jqe_A_D
[CQA]: 2aou_A_C
[CQA]: 2aou_A_C
[CQA]: 2aou_A_D
[CQA]: 2aou_A_D
[CQA]: 2aou_B_E
[CQA]: 2aou_B_E
[CQA]: 2aou_B_F 2aou_B_F
|
| 41 | SAP_HUMAN SAPOSIN-B | 78 |
[CLQ]: 4v2o_A_E
[CLQ]: 4v2o_A_E
[CLQ]: 4v2o_B_D
[CLQ]: 4v2o_B_D
[CLQ]: 4v2o_B_D
[CLQ]: 4v2o_B_D
[CLQ]: 4v2o_B_D
[CLQ]: 4v2o_B_D
[CLQ]: 4v2o_B_D
[CLQ]: 4v2o_B_D
[CLQ]: 4v2o_C_E 4v2o_C_E
|
| 42 | EIS_MYCTU N-acetyltransferase Eis | 392 |
[CLQ]: 8f4z_A_B
[CLQ]: 8f4z_A_B
[CLQ]: 8f4z_A_B
[CLQ]: 8f4z_A_B
[CLQ]: 8f4z_A_B
[CLQ]: 8f4z_A_B
[CLQ]: 8f4z_A_B
[CLQ]: 8f4z_A_B
[CLQ]: 8f4z_A_B
[CLQ]: 8f4z_A_B
[CLQ]: 8f4z_A_B 8f4z_A_B
|
| 43 | TYTR_TRYCR TRYPANOTHIONE REDUCTASE (OXIDIZED FORM) | 485 |
[QUM]: 1gxf_A_E
[QUM]: 1gxf_A_E
[QUM]: 1gxf_A_F
[QUM]: 1gxf_A_F
[QUM]: 1gxf_A_I
[QUM]: 1gxf_A_I
[QUM]: 1gxf_B_F
[QUM]: 1gxf_B_F
[QUM]: 1gxf_B_H
[QUM]: 1gxf_B_H
[QUM]: 1gxf_B_I 1gxf_B_I
|
| 44 | ADRB2_HUMAN:D9IEF7_BPT4:ADRB2_HUMAN Fusion protein of Beta-2 adrenergic receptor and T4 Lysozyme | 442 |
[SNP]: 6ps5_A_B
[CAU]: 6ps5_A_B
[CAU]: 2rh1_A_I
[CAU]: 2rh1_A_I
[CAU]: 5d5a_A_C
[CAU]: 5d5a_A_C
[CAU]: 5d5b_A_C
[CAU]: 5d5b_A_C
[CAU]: 5d6l_A_I
[CAU]: 5d6l_A_I
[CAU]: 5x7d_A_G
[CAU]: 5x7d_A_G
[CAU]: 6ps0_A_B 6ps0_A_B
|
| 45 | Q94LW9_9GENT Strictosidine synthase | 302 |
[KWK]: 6s5u_A_G
[KWK]: 6s5u_A_G
[KWK]: 6s5u_A_H
[KWK]: 6s5u_A_H
[KWK]: 6s5u_B_I
[KWK]: 6s5u_B_I
[KWK]: 6s5u_B_J
[KWK]: 6s5u_B_J
[KWK]: 6s5u_C_K
[KWK]: 6s5u_C_K
[KWK]: 6s5u_C_L
[KWK]: 6s5u_C_L
[KWK]: 6s5u_D_N
[KWK]: 6s5u_D_N
[KWK]: 6s5u_D_O
[KWK]: 6s5u_D_O
[KWK]: 6s5u_E_P
[KWK]: 6s5u_E_P
[KWK]: 6s5u_E_Q
[KWK]: 6s5u_E_Q
[KWK]: 6s5u_F_R
[KWK]: 6s5u_F_R
[KWK]: 6s5u_F_S 6s5u_F_S
|
| 46 | RXRA_HUMAN Retinoic acid receptor RXR-alpha | 214 |
[1O8]: 4k4j_A_C
[1O8]: 4k4j_A_C
[1O8]: 4k4j_A_C
[29V]: 4k4j_A_C
[29V]: 4m8e_A_C
[29V]: 4m8e_A_C
[29V]: 4m8e_A_C
[R4M]: 4m8e_A_C
[R4M]: 4m8h_A_C
[R4M]: 4m8h_A_C
[R4M]: 4m8h_A_C
[2VR]: 4m8h_A_C
[2VR]: 4poh_A_C
[2VR]: 4poh_A_C
[2VR]: 4poh_A_C
[2VP]: 4poh_A_C
[2VP]: 4poj_A_C
[2VP]: 4poj_A_C
[2VP]: 4poj_A_C
[2VZ]: 4poj_A_C
[2VZ]: 4pp3_A_C
[2VZ]: 4pp3_A_C
[2VZ]: 4pp3_A_C
[3RB]: 4pp3_A_C
[3RB]: 4rfw_A_C
[3RB]: 4rfw_A_C
[3RB]: 4rfw_A_C
[2W0]: 4rfw_A_C
[2W0]: 4pp5_A_C
[2W0]: 4pp5_A_C
[2W0]: 4pp5_A_C 4pp5_A_C
|
| 47 | NQO2_HUMAN Ribosyldihydronicotinamide dehydrogenase [quinone] | 230 |
[0TX]: 4fgk_A_D
[0TX]: 4fgk_A_D
[0TX]: 4fgk_A_F
[0TX]: 4fgk_A_F
[0TX]: 4fgk_B_D
[0TX]: 4fgk_B_D
[0TX]: 4fgk_B_F
[CLQ]: 4fgk_B_F
[CLQ]: 4fgl_A_G
[CLQ]: 4fgl_A_G
[CLQ]: 4fgl_A_J
[CLQ]: 4fgl_A_J
[CLQ]: 4fgl_B_G
[CLQ]: 4fgl_B_G
[CLQ]: 4fgl_B_J
[CLQ]: 4fgl_B_J
[CLQ]: 4fgl_C_M
[CLQ]: 4fgl_C_M
[CLQ]: 4fgl_C_P
[CLQ]: 4fgl_C_P
[CLQ]: 4fgl_D_M
[CLQ]: 4fgl_D_M
[CLQ]: 4fgl_D_P
[CLQ]: 4fgl_D_P
[CLQ]: 4fgl_D_Q
[1PQ]: 4fgl_D_Q
[1PQ]: 4fgj_A_E
[1PQ]: 4fgj_A_E
[1PQ]: 4fgj_A_F
[1PQ]: 4fgj_A_F
[1PQ]: 4fgj_B_E
[1PQ]: 4fgj_B_E
[1PQ]: 4fgj_B_F 4fgj_B_F
|